Playing around with CellNOptR Tool and MIDAS File
By Güngör Budak
- 3 minutes read - 427 wordsWith CellNOptR, we will try to construct network models for the challenge. For this, the tool needs two inputs. First one is a special data object called CNOlist that stores vectors and matrices of data. Second one is a .SIF file that contains prior knowledge network which can be obtained from pathway database and analysis tools.
CNOlist contains following fields: namesSignals, namesCues, namesStimuli and namesInhibitors, which are vectors storing the names of measurements. Another vector called timeSignals stores time points. The field valueSignals contains protein abundance measurements and the fi elds valueCues, valueStimuli and valueInhibitors) are boolean matrices with a 1 or 0, for each condition (row) when the corresponding cue (column) is present or absent, respectively [4].
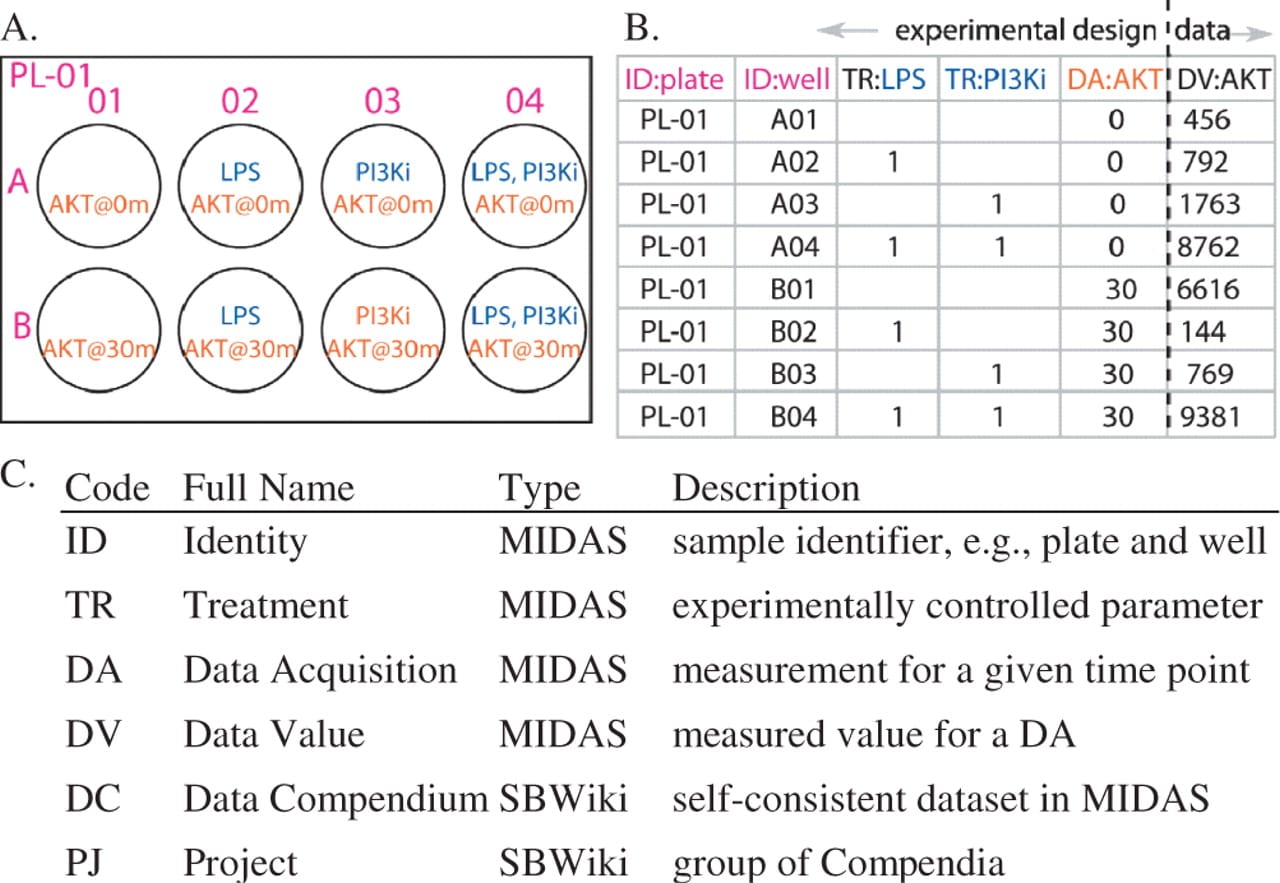
A. Experimental setup; B. Data obtained from the experiment; C. MIDAS description. Taken from Bioinformatics (2008) 24 (6): 840-847.doi: 10.1093/bioinformatics/btn018*
We are given MIDAS files and each row in a MIDAS file belongs to a single experimental sample and each column is one sample attribute, such as identity, treatment condition, or value obtained from an experimental assay. The column header has two values: a two-letter code defining the type of column, (e.g. TR for treatment, DV for data value), and a short column name (e.g. a small molecule inhibitor added or a protein assayed). Each cell stores the corresponding value for the row (sample) such as a presence or absence of stimulus/inhibitor, time point, or data value [1].
CellNOptR has a function that can read a MIDAS file and with another one it is possible to convert it to CNOlist and work on it. readMIDAS gets the content of CSV file to a data object and using that data object, makeCNOlist makes a CNOlist[2]. After I did it and look at the CNOlist, it worked but there were some issues with naming. Stimuli vector was named as inhibitors and inhibitors vector was named as stimuli. It also stores variances of data values. The variances are not zero if replicates are found [3].
So I was able to read experimental data as an object with a small problem in naming that can be solved easily by changing the names of the vectors.
This was the first input required for CellNOptR tool. Second one is the prior knowledge network and in the following days, I will work on it.
References
- Flexible informatics for linking experimental data to mathematical models via DataRail, http://bioinformatics.oxfordjournals.org/content/24/6/840.full
- Training of boolean logic models of signalling networks using prior knowledge networks and perturbation data with CellNOptR, http://www.bioconductor.org/packages/release/bioc/vignettes/CellNOptR/inst/doc/CellNOptR-vignette.pdf
- Package ‘CellNOptR’, http://www.bioconductor.org/packages/release/bioc/manuals/CellNOptR/man/CellNOptR.pdf
- In Silico Systems Biology: Scripting with CellNOptR, http://www.cellnopt.org/cytocopter/resources/tutorial_wtac_2013.pdf