Experimental Data Optimization for Network Inference
By Güngör Budak
- 2 minutes read - 291 wordsAs I mentioned in my previous post, experimental data from the challenge has missing data values that create problems during analyses. To solve it, first thing I did was to optimize data, which includes detecting missing conditions and putting NAs for data values and sorting them if necessary.
I wrote two functions in the script. First one ranks the data according to the fashion and sorts it based on these ranks. The second one gets sorted list and looks for missing data values and puts NAs as data values.
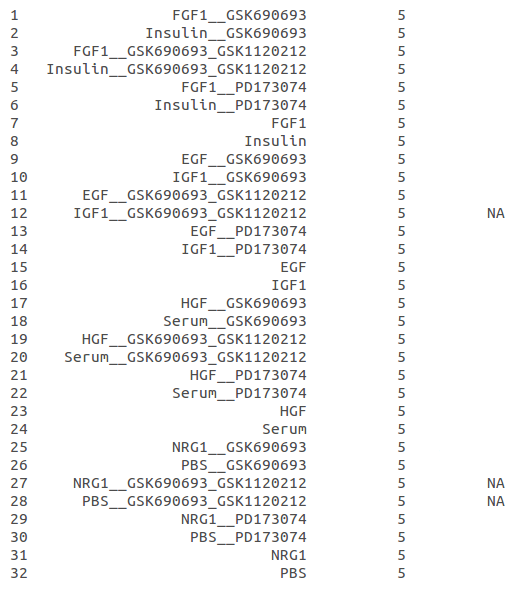
Above there is the list of conditions which are designed to represent “fashion” in the data. Also, it’s the result of the functions I mentioned above and for this time point, there are 3 missing data values. Actually, “NRG1__GSK690693_GSK1120212” and “PBS__GSK690693_GSK1120212” are missing for all time points. So I won’t be able to make any inference using these conditions. “IGF1__GSK690693_GSK1120212” is the only data value missing among the others. There are some other missing data values in different time points and all of these can be estimated.
The first approach I will use for the estimation is calculating formula for regression line and finding corresponding data value using time point. There are several others and I will mention them later.
But before moving on to estimation part, I want to be sure that this optimization approach is working for all cell lines. I applied this approach for MCF7 cell line and it wasn’t working as expected. Because in MCF7 cell line data DMSO treatments (no inhibitors) have duplicates, which cannot be handled by this version of script. This is also similar for UACC812 and BT549 cell line.
I will modify the script to work with these and then move to estimation part.